The Data Highlights section showcases recently published datasets relevant to pandemic preparedness research by researchers affiliated with a Swedish research institute. Specifically, in our case we write about e.g. COVID-19 and SARS-CoV-2, infectious diseases in general, and antibiotic resistance. The goal is to highlight openly shared data that can potentially be used by many other researchers to make further discoveries.
To easily find data highlights relevant to a specific topic, simply click on the coloured tag and the page will filter and display only the highlights related to that topic. Alternatively, use the search bar to find highlights of potential interest.If you have published or are about to publish data that you think should be highlighted in this section, please click the button below to suggest a data highlight and the editorial team will get in touch with you as soon as possible to discuss your suggestion.
May 21, 2024
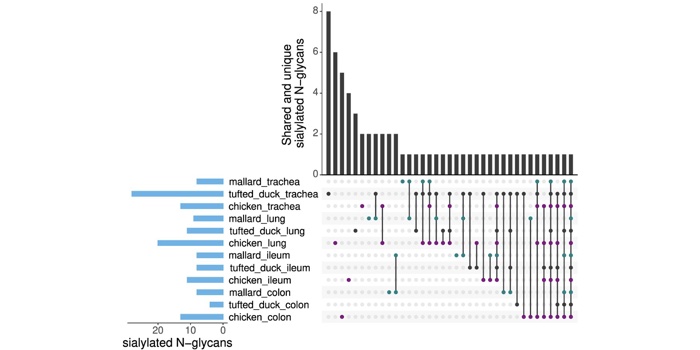
New findings from Nilsson and colleagues showed that sialylated glycan structures, previously thought to be exclusive to mammals, are present in the three avian species studied.
January 19, 2024
Babačić and colleagues expanded the coverage of the soluble blood proteome using mass spectrometry. In order to support further research in this area, their results have been added to an open-access app.
September 20, 2023
This study by Knöppel, Broström et al is a large effort to elucidate replication initiation in bacteria. The authors have openly shared over 3 TB of microscopy imaging data.
June 2, 2023
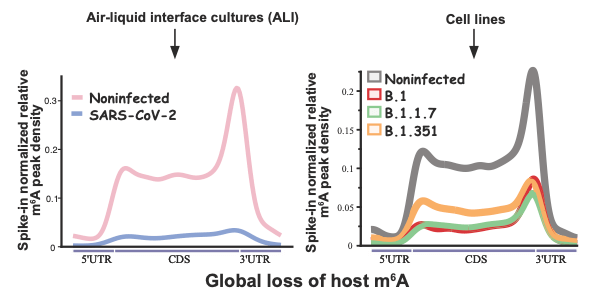
Vaid and Mendez, and collaborators, studied how the gene expression profile of m6A mRNA is affected both during and after COVID-19 infection. All sequencing data and the source code for analysis are shared.
May 5, 2023
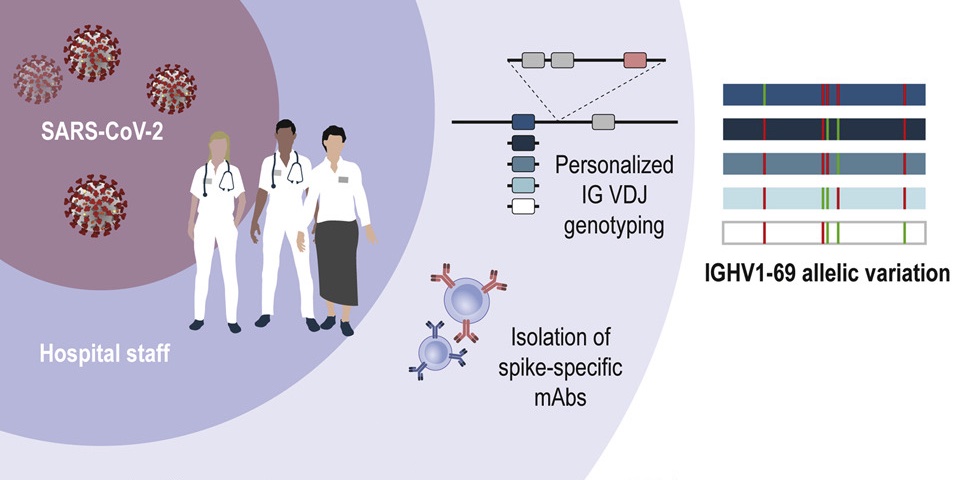
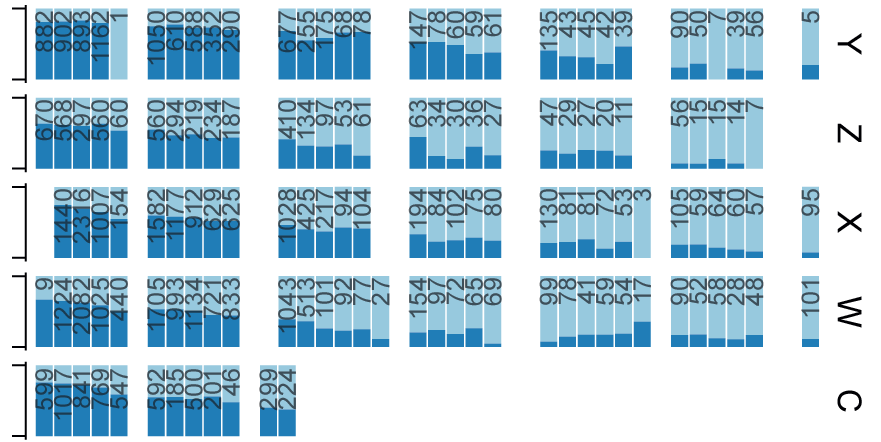
Pushparaj and colleagues use genotyping and haplotype analysis to show high genetic diversity in IGH genes among humans, which may influence our response to infections. Data, and IgDiscover software shared.
April 18, 2023
A study from Elf lab at Uppsala University/SciLifeLab shows perseverance can be a reason for antibiotic resistance development in E. coli. Microscopy image data shared in SciLifeLab Data Repository.
December 19, 2022
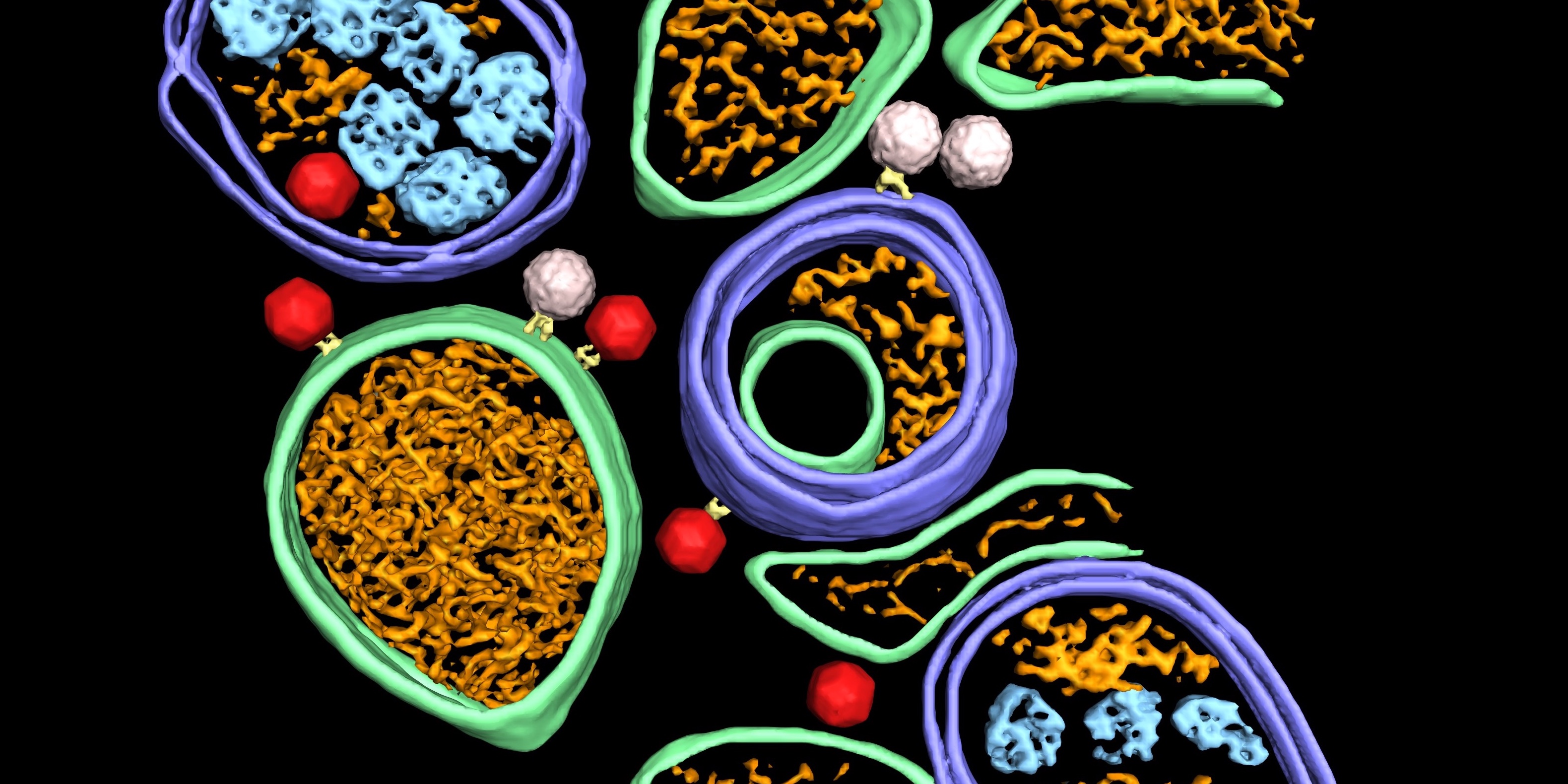
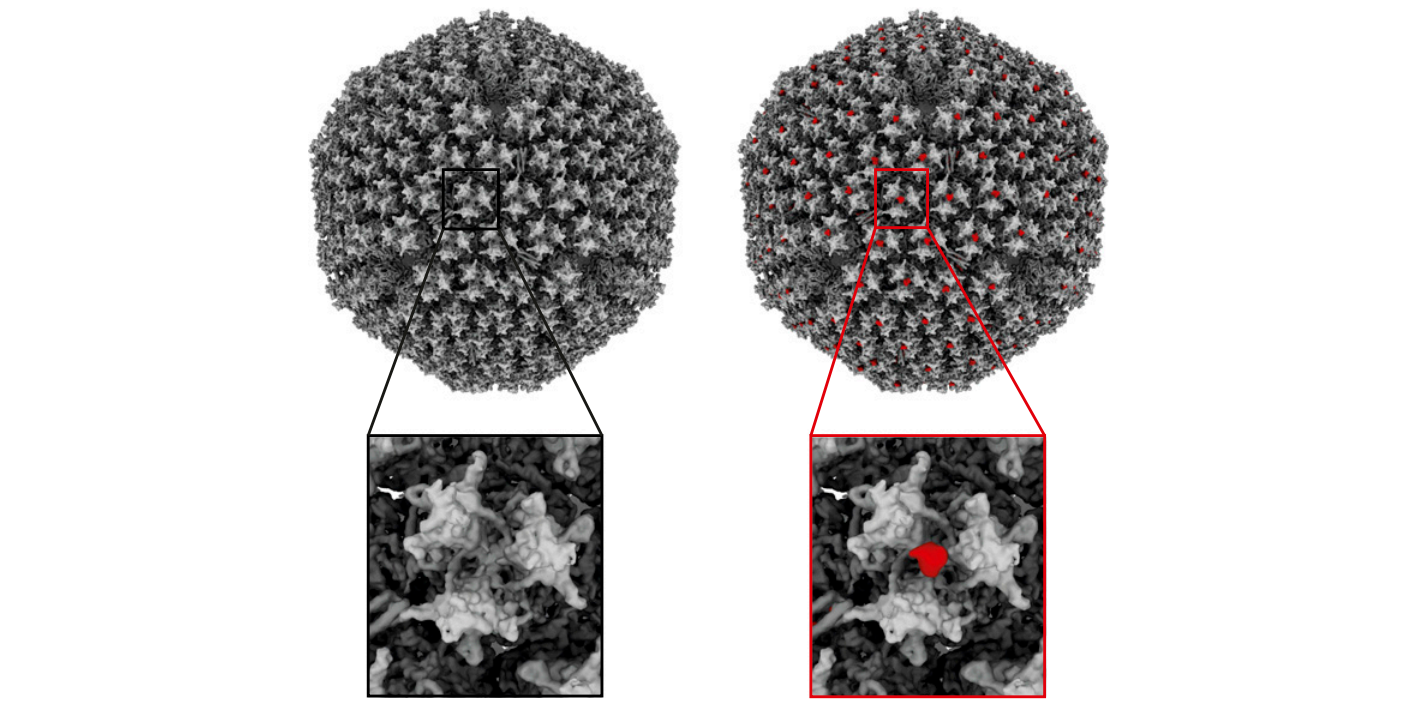
Dahmane et al (2022) used Cryo-electron tomography to provide an integrated structural framework for multiple stages of the poliovirus life cycle. Data and code are shared openly.
November 22, 2022
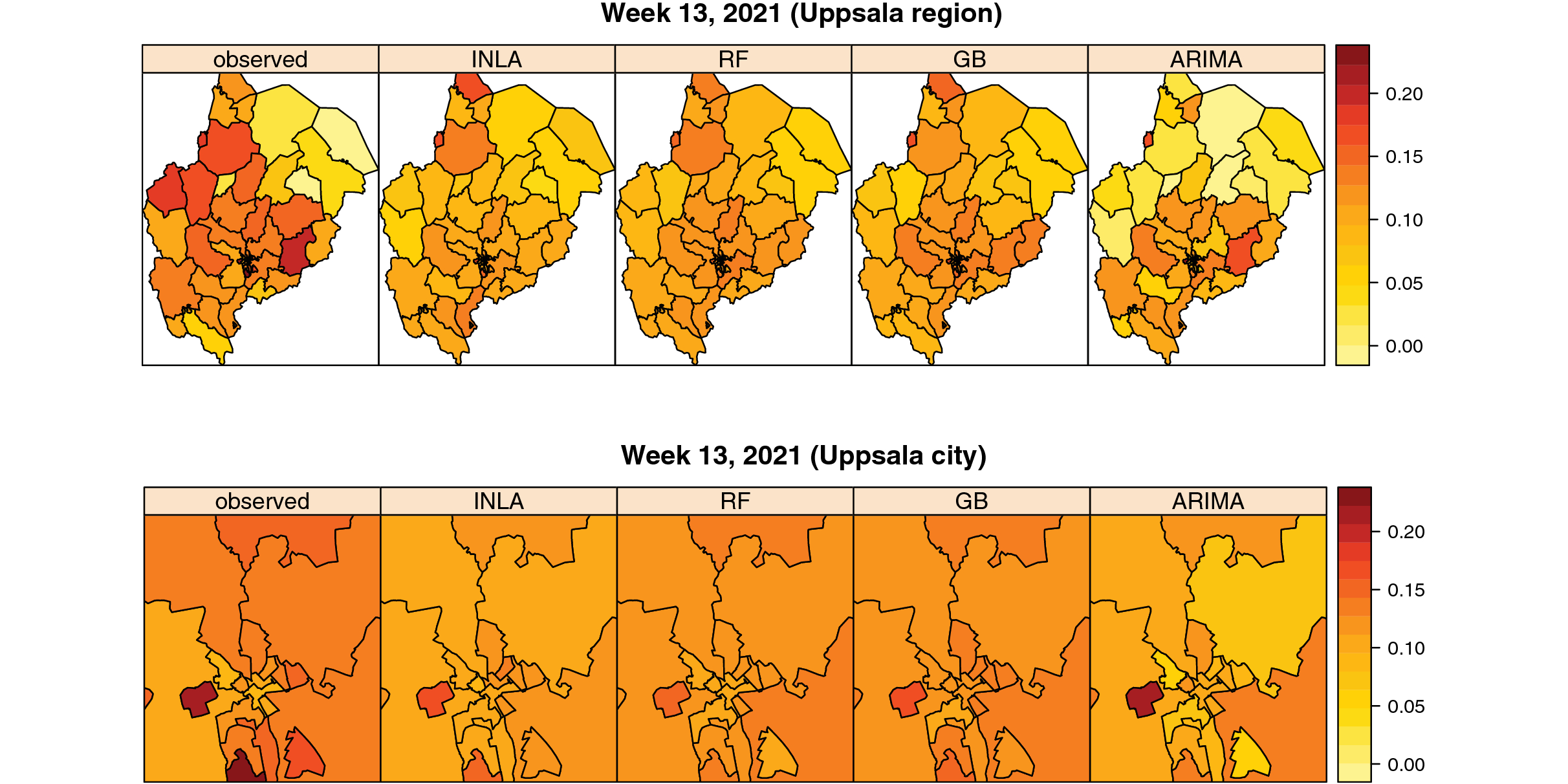
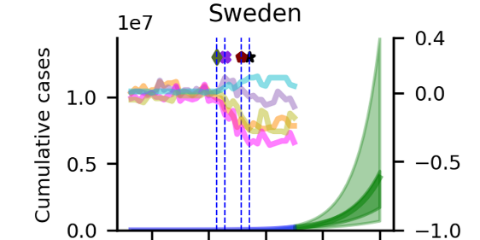
Van Zoest et al (2022) developed and evaluated the performance of four different statistical single models and one ensemble model to predict trends in COVID-19 test positivity. Data are shared openly in GitHub.
October 3, 2022
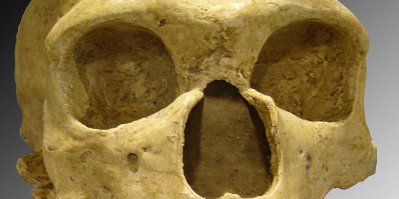
Study by Hugo Zeberg and Svante Pääbo fully based on openly available data published open access in Nature.
Now a Nobel Prize winner!!
Now a Nobel Prize winner!!
August 15, 2022
Lentini and collegues used direct RT-PCR to show the transition from Omicron BA.1 to sub-variant BA.2 in Sweden Jan-March 2022. New Preprint from Reinius lab which shares data and code.
May 6, 2022
New study gives insight into host-viral interactions of Crimean-Congo hemorrhagic fever, an infectious disease without available treatments. Raw RNAseq, mass spectrometry proteomics data and code shared.
April 25, 2022
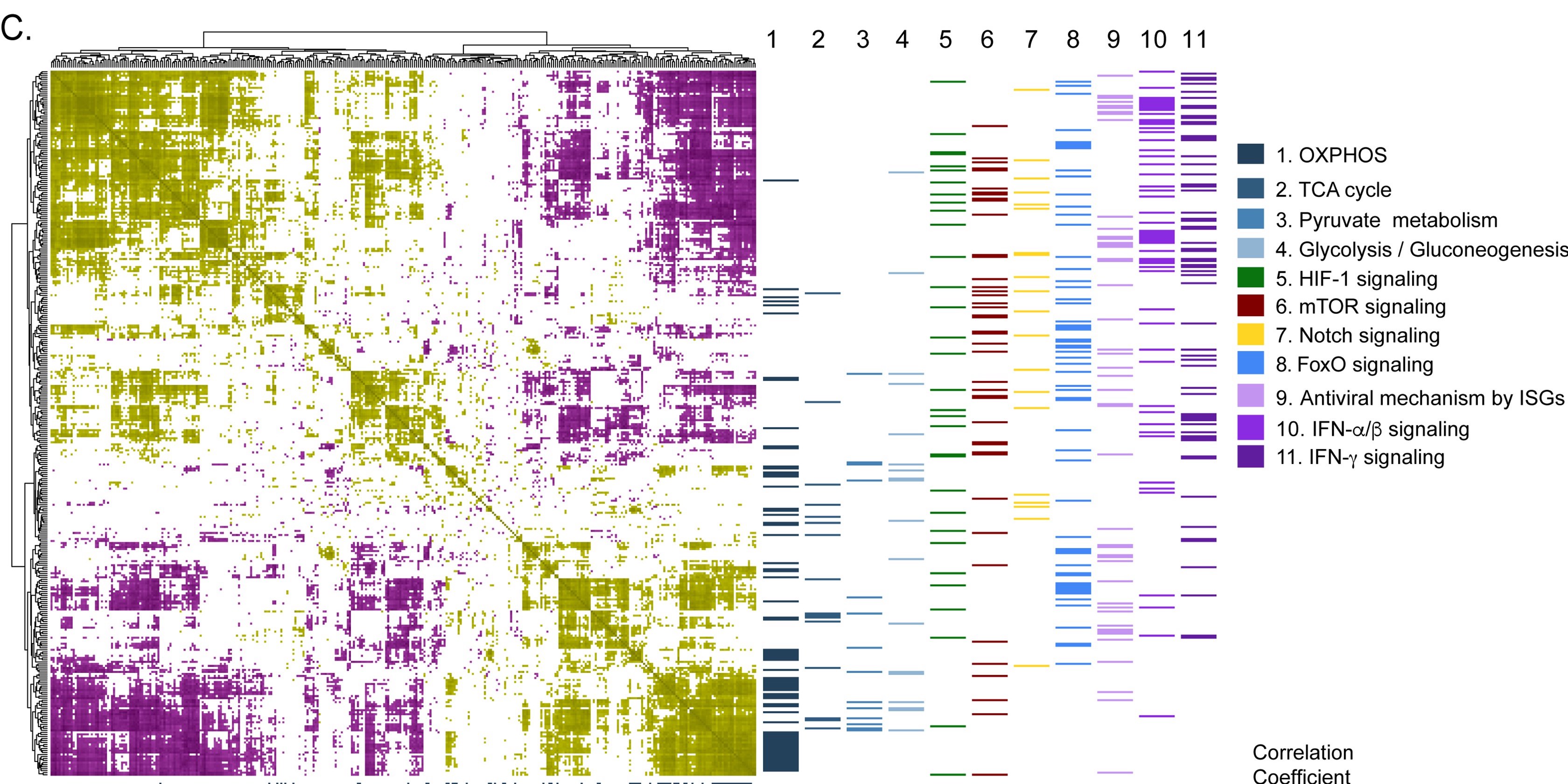
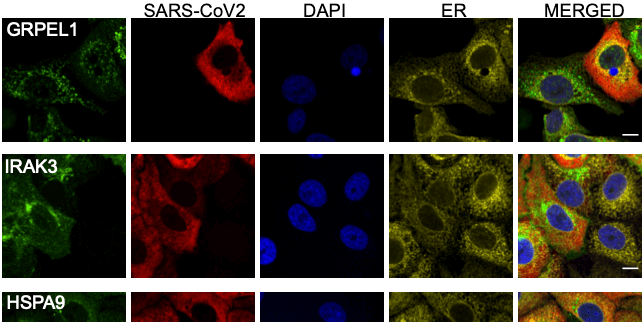
Expression of 602 host proteins were evaluated in populations of infected and non-infected cells using immunofluorescence. ≈75,000 images have been published as a resource for further studies.
March 1, 2022
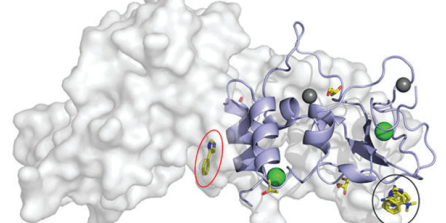
Interactions between fragments and SARS-CoV-2 nsp10 have been successfully identified and characterised by Kozielski and colleagues. Data shared on the Protein Data Bank and Zenodo.
February 21, 2022
Mangsbo and colleagues investigated sensitivity and specificity of a SARS-CoV-2 T-cell assay. Research warrants further research into that future T-cell receptor sequencing as future tools for prediction of previous infections.
February 17, 2022
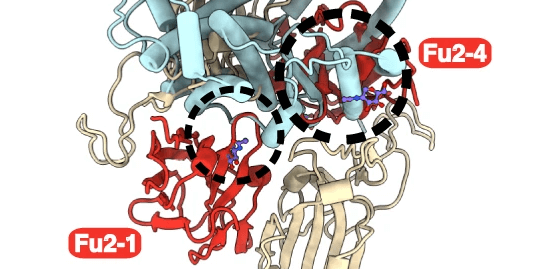
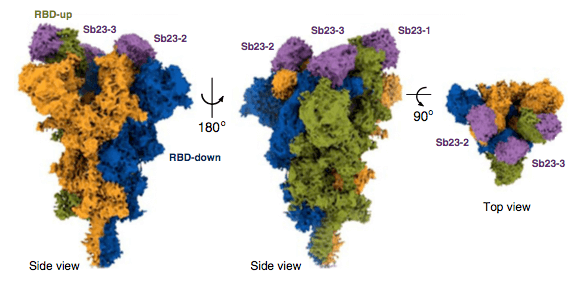
In this study, Hanke and colleagues discovered a new nanobody. The new RBD-specific Fu2 nanobody was found to utilize a unique neutralization mechanism. Data was shared openly in, eg. The Electron Microscopy Data Bank.
February 9, 2022
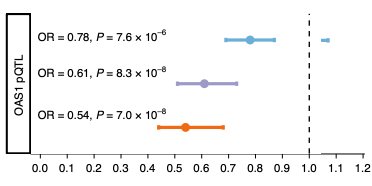
In this article, Huffman et al studied individuals across different ancestry groups and discusses the association between OAS1 splicing and severity of COVID-19. Data related to this study is shared publicly.
January 20, 2022
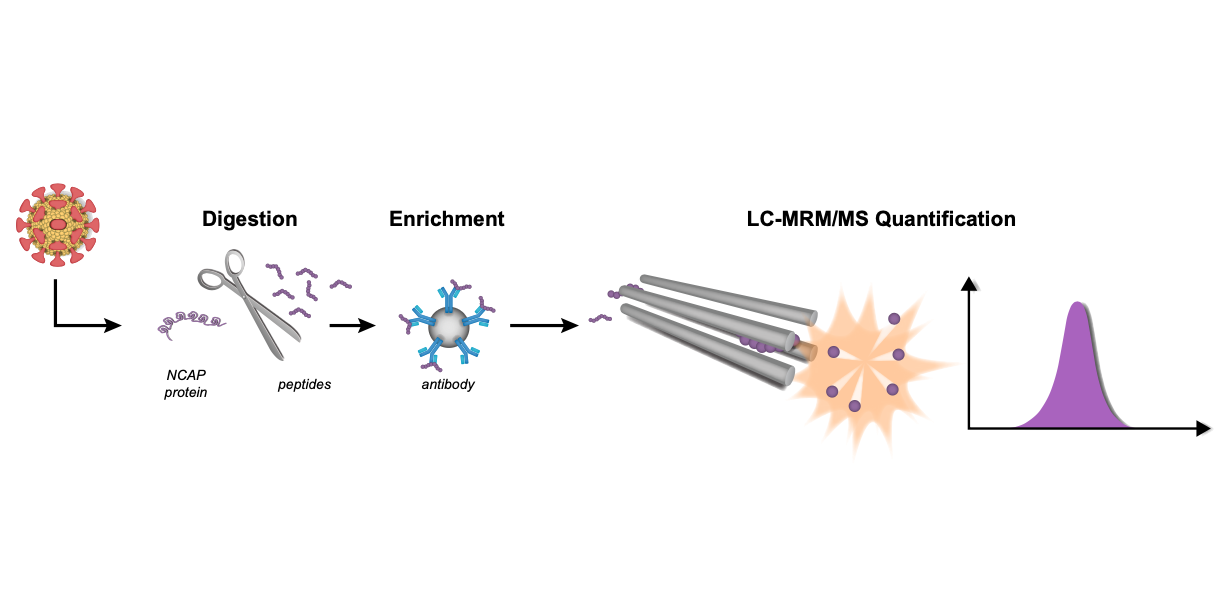
Hober et al. present how LC-MRM/MS could form the basis of a scalable method that should facilitate future pandemic preparedness. The data has been openly shared.
November 24, 2021
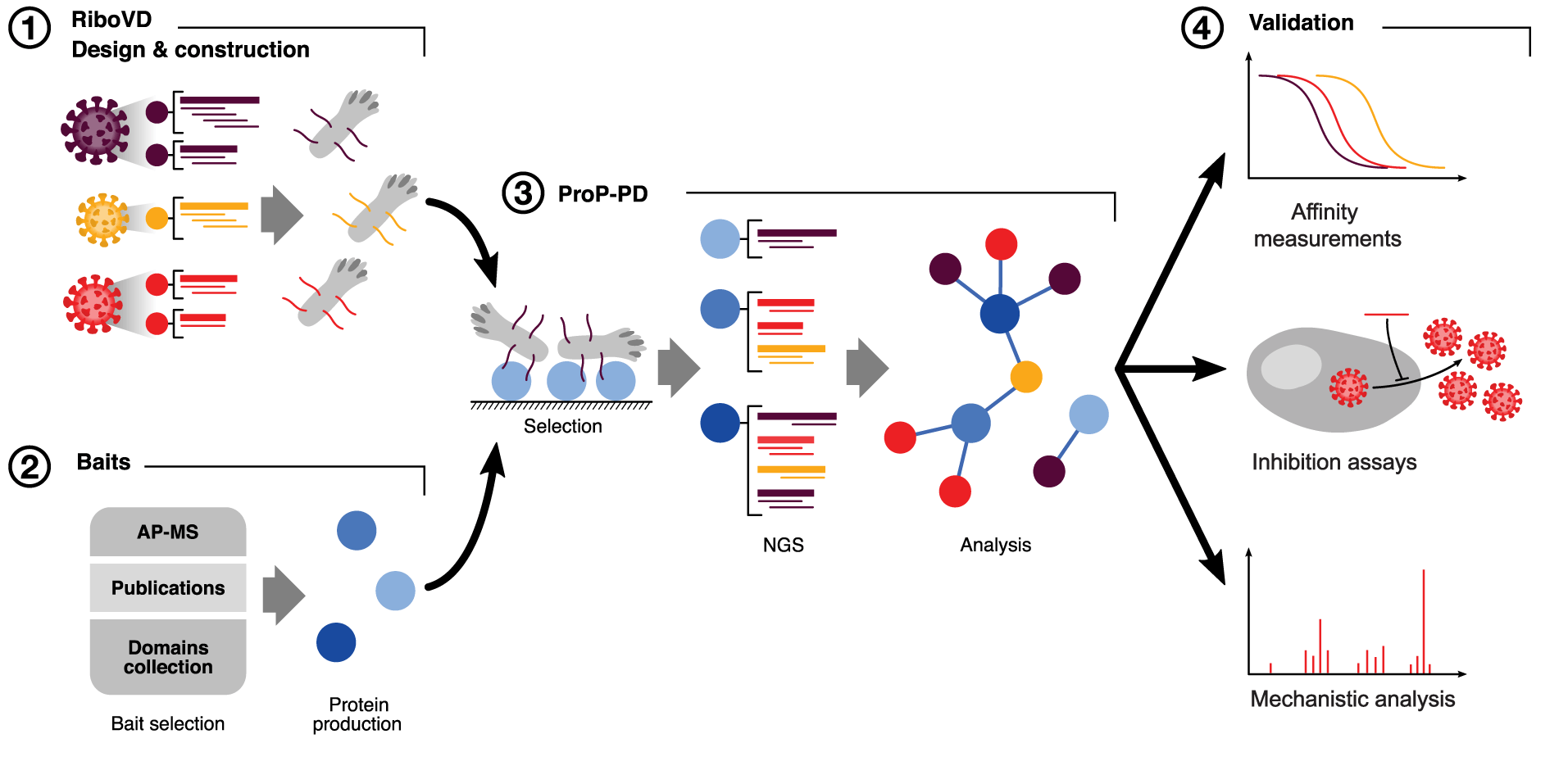
Kruse and Benz et al. describe a scalable method of investigating virus-host interactions that can be used as a means of identifying potential targets for novel antiviral drugs. All data from the study have been shared publicly.
November 19, 2021
Recent study finds that drug repurposing can offer an average of an 11-fold increase in disease coverage and demonstrates its value for COVID-19. All data and code that support the findings have been made publicly available.
November 3, 2021
The COVID Human Genetic Efforts consortium published a study on genetic factors underlying greater susceptibility to the severe COVID-19 in men. Relevant RNA-seq data were shared in Gene Expression Omnibus (GEO).
October 18, 2021
Roxhed et al. demonstrate how self sampling at home can be effective by profiling IgG and IgM levels against several versions of SARS-CoV-2 proteins (S, RBD, N) in home-sampled dried blood spots. Metadata records and the analysis scripts used in the study are shared.
October 7, 2021
Krishnan et al. showed that the SARS-CoV-2 virus can use and rewire metabolic pathways to their benefit in replication. Their findings indicate that host metabolic perturbation could be an attractive strategy to limit replication. Code and data from the study have been shared pubicly.
August 10, 2021
Researchers in the COVID-19 Host Genetics initiative performed genome-wide association meta-analyses to examine the loci involved in the variation of COVID-19 disease severity and susceptibility between individuals. The data and code used in the project have been made available.
August 2, 2021
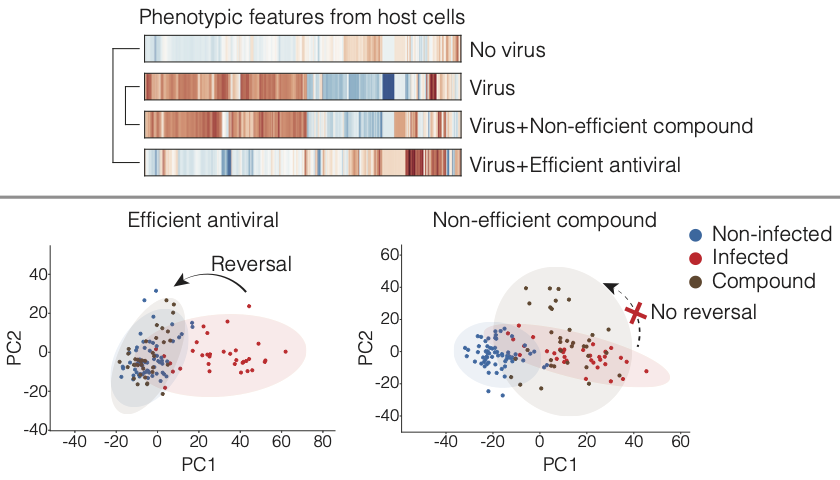
Rietdijk and colleagues used a new phenomics approach to aid in the discovery of both novel drugs and drugs that could potentially be repurposed to fight COVID-19. Both data and code are published.
July 6, 2021
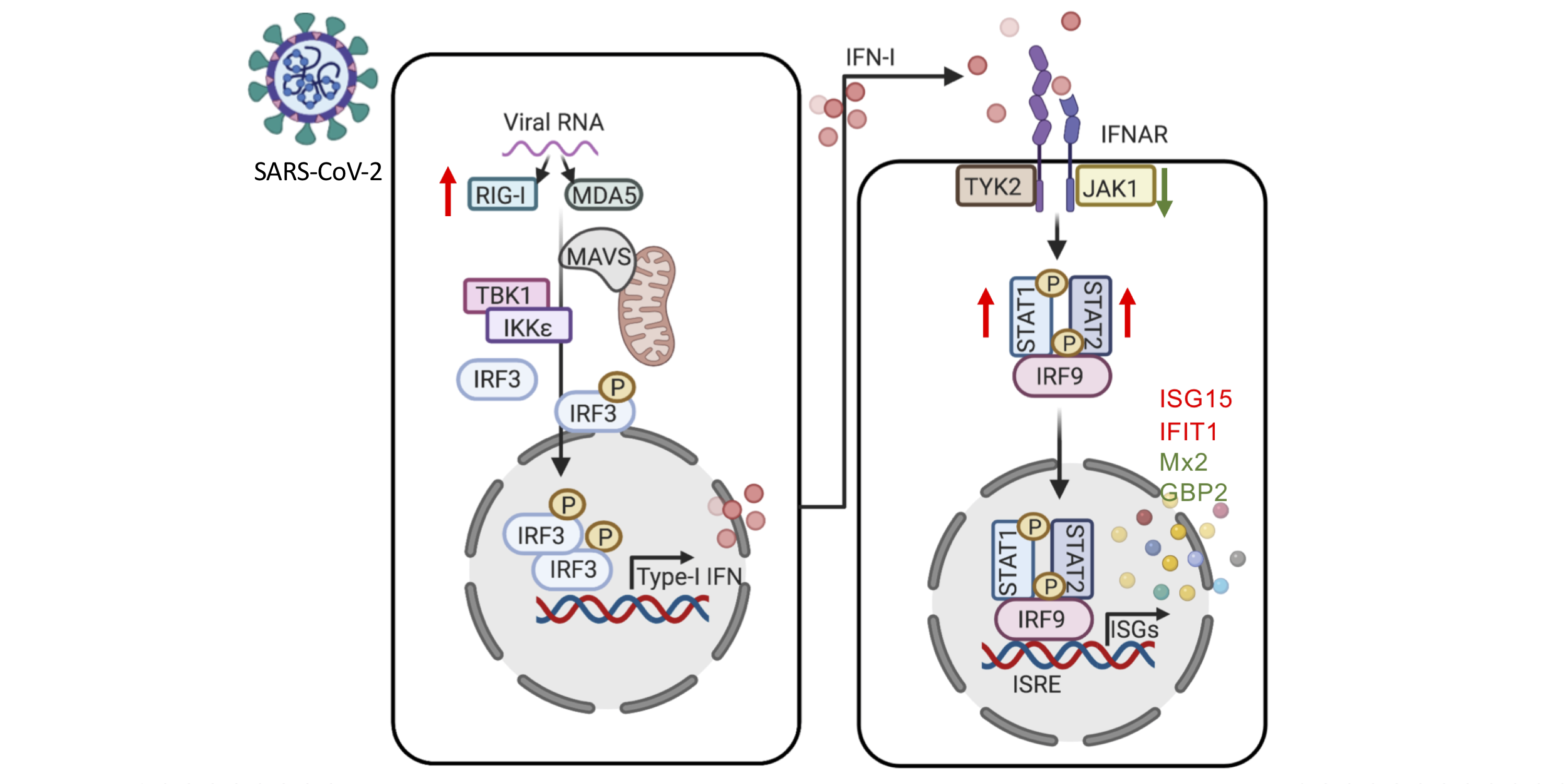
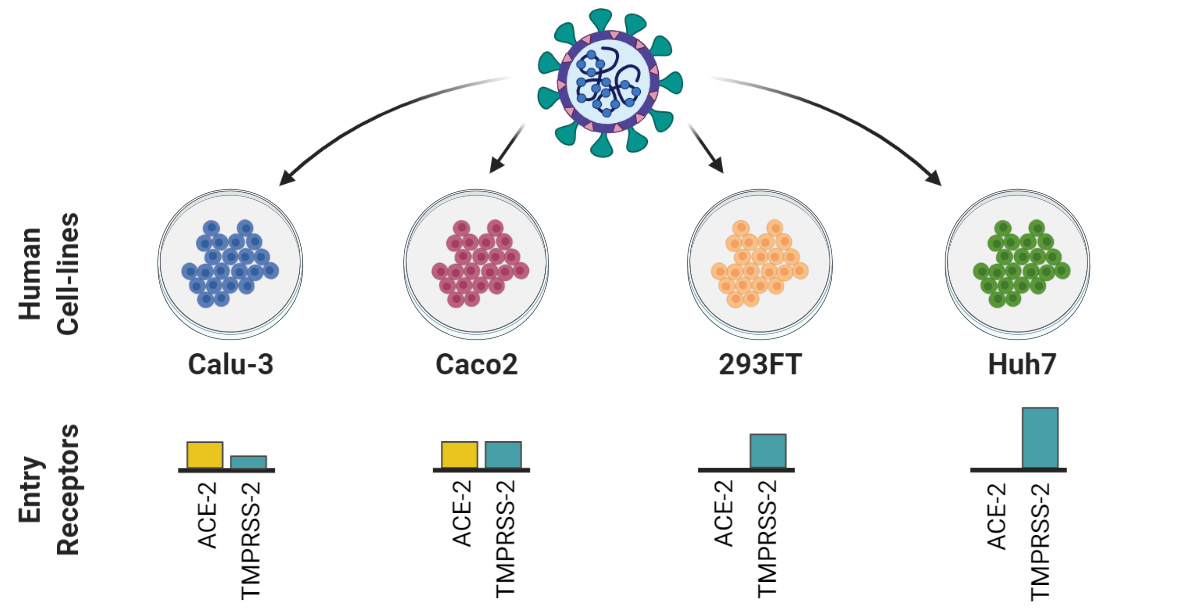
Chen and Saccon et al. used a proteomics-based approach to study how SARS-CoV-2 infection affected Type-1 interferon signalling in liver cells. Both data and code are published.
July 1, 2021
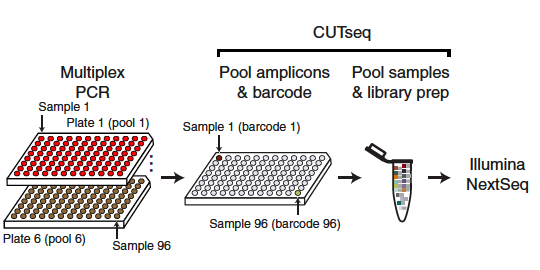
Simonetti, Zhang, and Harbers et al. have shared a cost-effective, scalable and versatile method (alongside associated data and code) that could facilitate large-scale genomic surveillance of variants of concern (VOCs).
May 19, 2021
Saccon and colleagues studied changes in protein abundance caused by SARS-CoV-2 virus in susceptible cell lines using quantitative proteomics. All proteomics were made data openly available in different repositories.
April 13, 2021
Recent study examined a large number of inflammatory, immune response, cardiovascular, and neurological markers in the blood of patients variously impacted by COVID-19. Data and analysis code were shared in public repositories.
March 29, 2021
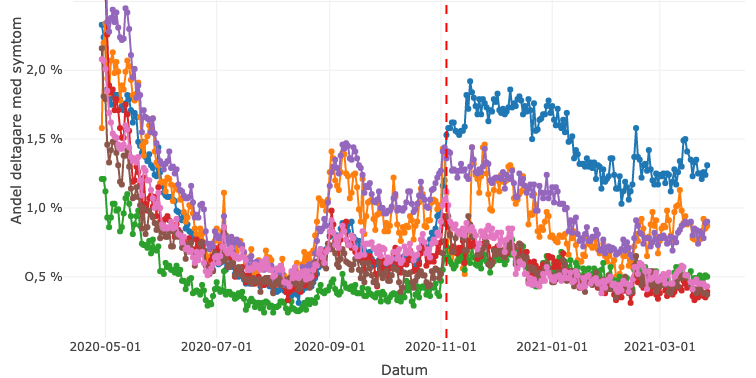
COVID Symptom Study Sweden collects data through a smartphone app to investigate prevalence, risk factors, and symptoms associated with COVID-19. To date, over 200.000 volunteers have enrolled in the study.
March 10, 2021
A new study largely relying on existing data re-use identified circulating proteins influencing COVID-19 susceptibility and severity. Based on these results, work on repurposing relevant existing drugs work can now be launched.
February 18, 2021
A recently published clinical study investigating bevacizumab, a humanized anti-VEGF monoclonal antibody, offers promising results in fighting severe COVID-19. Treatment progress data and analysis code were shared publicly.
February 16, 2021
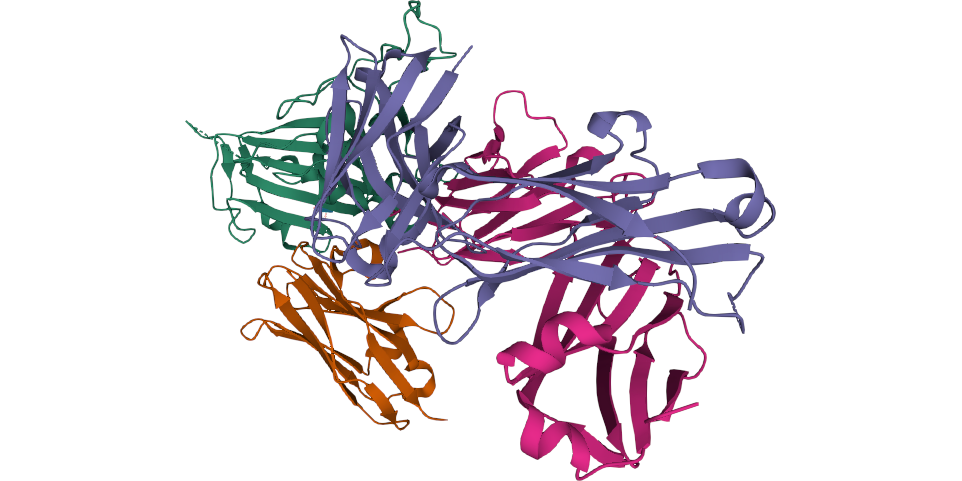
A recently published study characterizes the mechanisms by which nanobodies neutralize or prevent of infection in molecular detail. Crystallographic coordinates, cryo-EM density maps, and atomic coordinates were deposited to public databases.
January 18, 2021
New study identifies a noncanonical entry mechanism used by human adenoviruses, important for development of adenovirus-based vaccine vectors. The reported X-ray structural data was made available through the Protein Data Bank.
December 18, 2020
Newly discovered antiviral compounds showed broad-range antiviral activity against pathogenic RNA viruses such as SARS-CoV-2. The authors share data freely in open repositories.
December 7, 2020
Multiple autoantibodies and specific cytokines could be involved in the pathogenesis of MIS-C associated with COVID-19. Data and code made available in open repositories.
November 27, 2020
Leo Hanke and colleagues identified a SARS-CoV-2 RBD-specific single domain antibody fragment, Ty1, that potently neutralizes the virus. Ty1 should now be further investigated as a candidate for antiviral therapy.
November 24, 2020
Recently published study demonstrates that it is possible to select highly specific binders with neutralizing activity against SARS-CoV-2 from a synthetic nanobody library in a short timeframe.
November 18, 2020
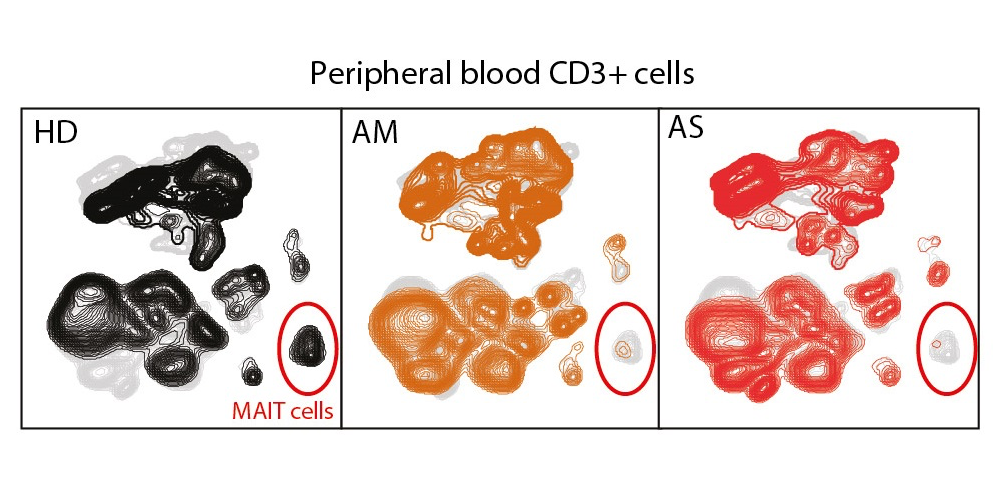
Tiphaine Parrot and colleagues studied changes in conventional and unconventional T cell subsets in patients with various disease severity using flow cytometry. The results were published along with the data.
November 3, 2020
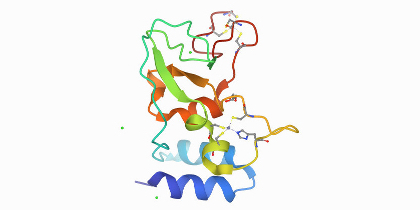
A study by Rogstam and colleagues published along with the crystal structure of SARS CoV-2 non-structural protein 10 in the Protein Data Bank provides a starting point for structure-guided drug discovery and design.
October 23, 2020
Patrick Bryant and Arne Elofsson shared code and instructions for modelling COVID-19 development using MCMC simulations based on mobile phone mobility data from Google mobility reports.
October 15, 2020
First study using data collected as part of the large-scale serology testing at SciLifeLab published
This study used only a fraction of the large volume of serology data continuously collected by the ScilifeLab Autoimmunity and Serology profiling facility.
October 1, 2020
Study by Björn Reinius and co-workers published open access in Nature Communications along with source data and the computational code.