Mechanism of entry used by human species D adenoviruses proving important for COVID-19 vaccine development
Published: 2021-01-18
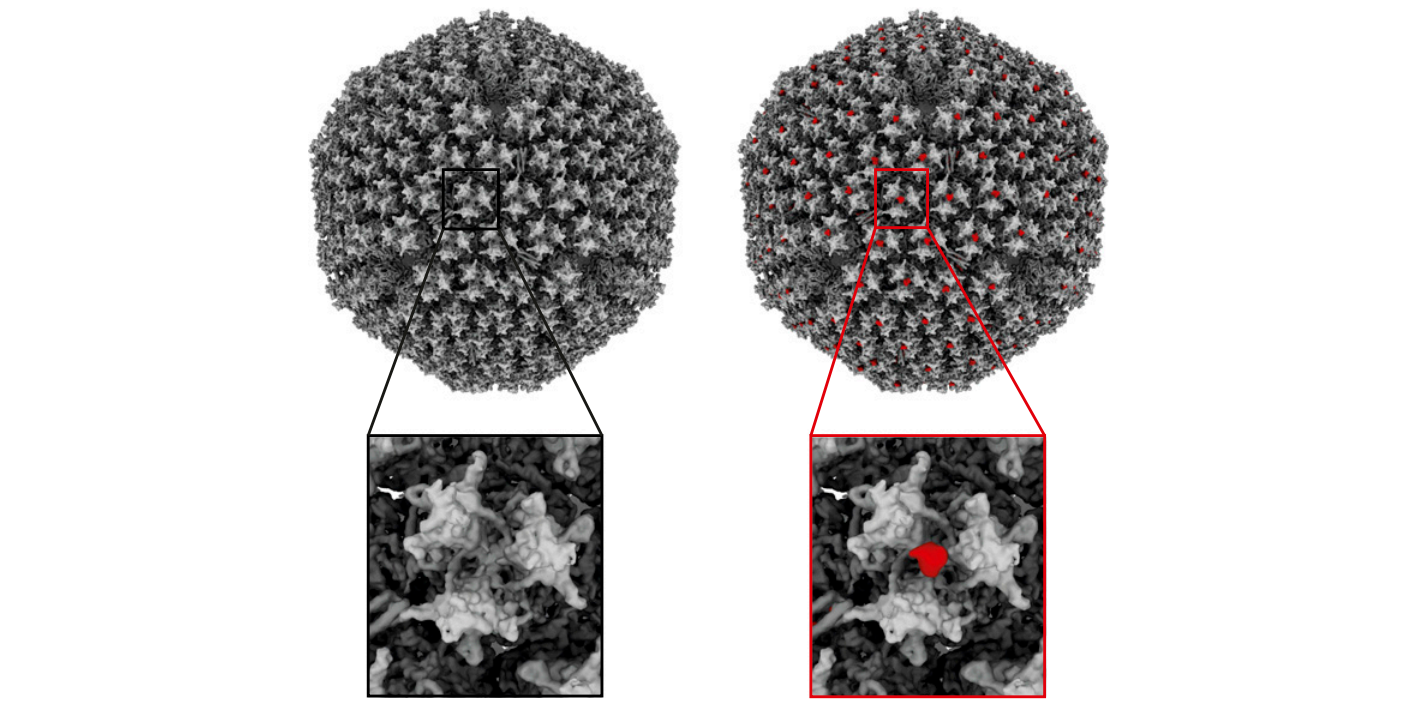
The COVID-19 pandemic has profoundly affected and changed society over the last year. Currently, in record time, a number of vaccines are under development. To date, two mRNA vaccines (Pfizer/BioNTech and Moderna) are in use in Sweden. Several other of the novel vaccines under development are based on genetically modified adenovirus vectors.
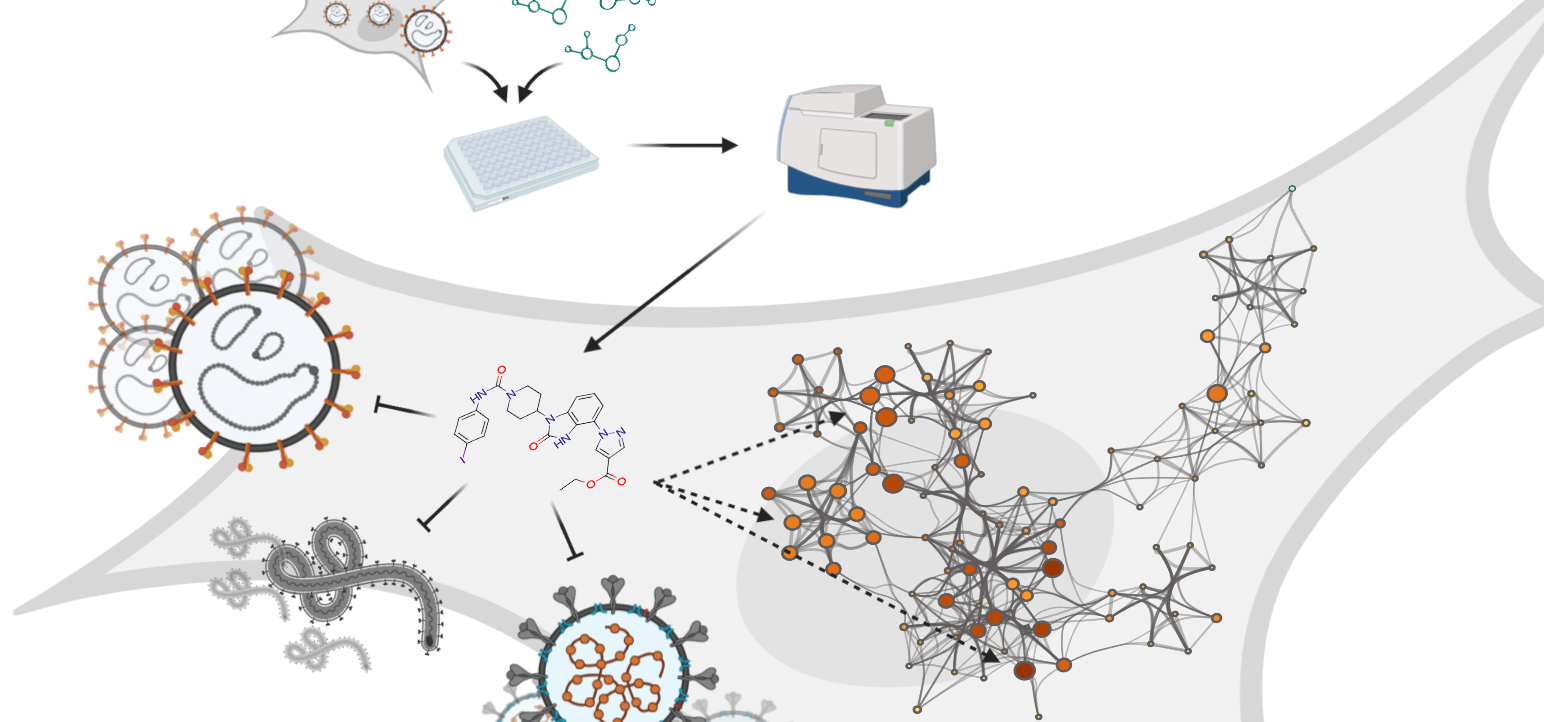
To date, over 100 human adenovirus (HAdV) types have been identified. The HAdV types are classified into seven species, named A-G. The HAdV species D (HAdV-D) comprises two-thirds of all known types and exhibit attractive feature as vaccine vectors. Research has shown that wild-type HAdVs are associated with different clinical manifestations in respiratory, lymphoid, gastrointestinal, and ocular tissues. The HAdV-D types are currently being explored as possible vaccine vectors for protection against COVID-19. However, the efficacy of vector-based vaccines depends on functional interactions with receptors on host cells. The adenovirus capsid protein is built by three main capsomers: hexon, fiber, and penton base. According to state-of-the-art, the fiber proteins anchor the virus to host cell receptors, followed by penton base proteins engaging co-receptors, which results in entry.
A recently published study led by B. David Persson and colleagues (PI: Niklas Arnberg), researchers from Umeå University, used a cell-based receptor-screening assay to identify CD46 as a receptor for human adenovirus species D56 (HAdV-D56). In addition, the researchers also validated the function of CD46 both in infection experiments using cells lacking and overexpressing CD46, and by competition infection experiments using soluble CD46. The results showed a difference between the human adenovirus species B and D types. HAdV-B types are known to engage CD46 through canonical interactions medidiated by the knob domain of the fiber protein, while HAdV-D types infect host cells through a direct interaction between CD46 and the hexon protein. Persson and colleagues showed that soluble hexon proteins inhibited HAdV-D56 infection, and that CD46 binds to HAdV-D56 and D26 hexons. Further, Persson and colleagues used cryo-EM analysis to solve the HAdV-D56 virion structure in the absence and presence of CD46, to 4.8 and 5.1 Å, respectively, and confirmed the interaction with the hexon protein. Interestingly, soluble CD46 was found to inhibit infection by 16 out of 17 investigated HAdV-D types, which may suggest CD46 as an important receptor for adenoviruses.
In summary, this study adds valuable information to our knowledge about adenovirus biology, which is of importance for development of adenovirus-based vaccine vectors used for the ongoing COVID-19 vaccine development to tackle the COVID-19 pandemic. In addition, HAdV-D types have been proposed a future vaccine vectors for other infections, i.e., HIV, respiratory syncytial virus, Ebola virus, and Zika virus.
The authors have deposited the X-ray structural data reported in the paper in the Protein Data Bank (7AJP, Crystal Structure of Human Adenovirus 56 Fiber Knob).
The Cryo-EM data were collected at the Umeå Core Facility for Electron Microscopy (SciLifeLab National Cryo-EM Facility and part of National Microscopy Infrastructure; NMI VR-RFI 2016-00968). This project has received funding from the Seventh Framework Programme; Marie-Curie Actions (324325 AD-VEC) (to N.A.), Human Frontier Science Program (CDA00047/2017-C) and Knut och Alice Wallenbergs Stiftelse (Wallenberg Centre for Molecular Medicine Fellowship) (to L.-A.C.), Stiftelsen Olle Engkvist Byggmästare (postdoctoral fellowship) (to K.R.), and Baden-Wurttemberg Foundation (M.S.).
Article
Persson, B. D., John, L., Rafie, K. Strebl, M. Frängsmyr, L. Ballmann, M. Z. Mindler, K. Havenga, M. Lemckert, A. Stehle, T. Carlson, L.-A, & Arnberg, N. Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46. Proceedings of the National Academy of Sciences, 118 (3) (2021).