Mass spectrometry suggested as a future essential analytical contribution to Covid-19 pandemic readiness
Published: 2022-01-20
Before entering the endemic phase of the COVID-19 pandemic, when the decision is taken to remove non-pharmaceutical interventions (NPIs) (i.e. restrictions for public gatherings, events and travel) and re-open societies, methods for population-wide/outbreak screenings must be put in place. Rapid and cost-effective specific molecular diagnostic tools for SARS-CoV-2 monitoring are therefore warranted. Such methods will, together with vaccination, treatments, and public awareness, be key for future pandemic readiness.
Real-time polymerase chain reaction (RT-PCR) is currently the technology most widely used for the detection of SARS-CoV-2. RT-PCR is also considered to be the “gold standard” by the World Health Organisation (WHO). RT-PCR tests have high analytical sensitivity and specificity. However, they do have some biases; positivity can linger over time, and they have relatively high false-positive rates. The false-positive rate depends on the kit and techniques used. Surkova et al. 2020 proposed that the rate of false-positives could be as high as 4% at certain test facilities. In addition, over the last few years, the shortage of diagnostic reagents (driven by the rapid buildup of large-scale laboratory facilities for SARS-CoV-2 detection) has caused a bottleneck for testing. Antigen tests have often been used as an alternative, as a result. Although antigen tests often exhibit similar specificity to PCR-based assays, they lack in sensitivity. The main advantage of the antigen tests is the simplicity of a binary read-out, as well as the possibility for self-testing and large-scale screening efforts.
For future pandemic readiness, recent studies have therefore suggested the use of liquid chromatography (LC) coupled with multiple reaction monitoring (MRM) tandem mass spectrometry (MS) detection for SARS-CoV-2. Using LC-MRM/MS to detect SARS-CoV-2 has several advantages. Firstly, the instrumentation can reliably quantify absolute protein concentrations, and secondly, the present-day MS instrumentation offers specificity, high precision, and high analytical sensitivity.
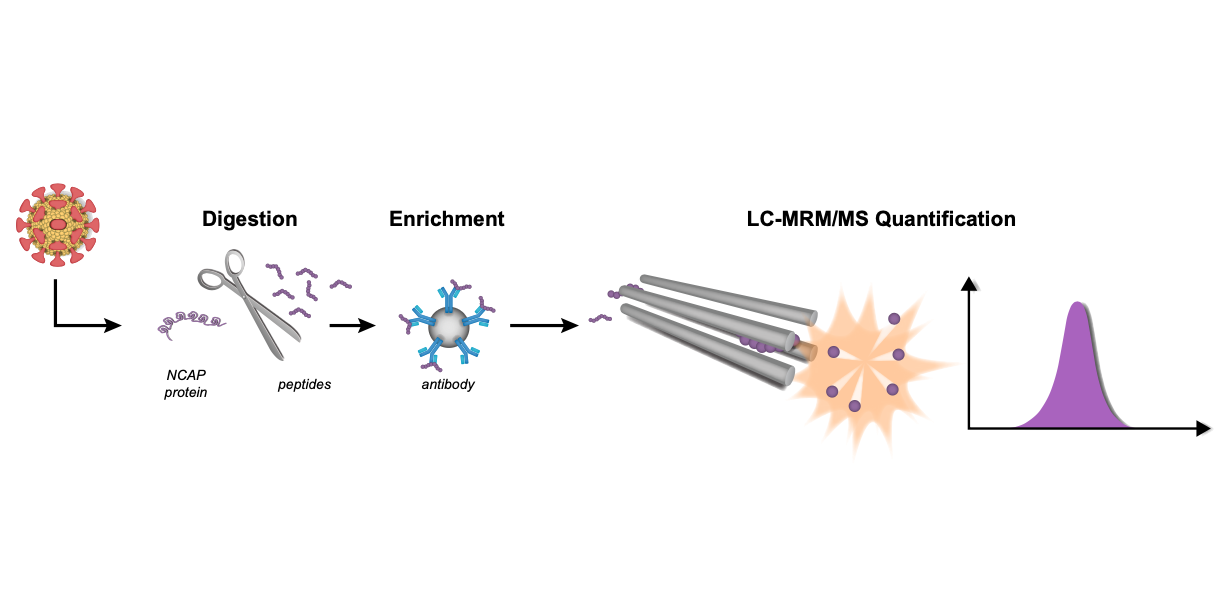
In a recently published article in eLife, researchers from Sweden, Canada, UK, and the Waters company (first author Andreas Hober (Science for Life Laboratory, The Royal Institute of Technology) and corresponding author Fredrik Edfors (Science for Life Laboratory, The Royal Institute of Technology)) describe the development of an analytical approach that is able to detect viral proteins based on peptide immuno-affinity enrichment combined with LC-MRM/MS.
Using a multiplexed strategy, Hober and colleagues used targeted proteomics analysis, in combination with read-out by LC-MRM/MS, to quantify and confirm the presence of SARS-CoV-2 in throat/nasopharynx/saliva samples. The samples used were self-collected from asymptomatic individuals as part of a workplace screen. The researchers performed RT-PCR, protein extraction and digestion, peptide enrichment, and LC-MRM/MS analysis as part of the study. The set-ups for the LC-MRM/MS used by Hober et al are, in brief, ACQUITY UPLC I-Class FTN system, with Binary Solvent Manager and column heater followed by a Xevo TQ-XS tandem MS (both from (Waters Corporation). The column was an ACQUITY Premier Peptide BEH C18 (2.1 mm × 50 mm, 1.7 µm, 300 Å column, Waters Corporation) and eluted at 0.6 ml/min. The mobile phases used were A (H2O/formic acid 0,1%) and B (CH3CN/formic acid 0.1%). A gradient was used for the run, followed by positive electrospray (ESI+;capillary voltage 0.5 k; source temperature 150°C; desolvation temperature 600°C, cone gas flow 150 l/h, and desolvation gas flow 1000 l/h). Finally, a TargetLynx XS (Waters Corporation) was used for the processing of the raw LC-MRM/MS data.
The results showed a good correlation between the levels of SARS-CoV-2 measured by LC-MRM/MS and the corresponding RT-PCR readout (r = 0.79). In addition, the analytical workflow had a similar turnaround time compared to regular RT-PCR instrumentation. The quantitative readout of viral proteins from the LC-MRM/MS corresponds to cycle thresholds Ct = 21-34. The proposed LC-MRM/MS-based method also has 100% negative percent agreement (estimated specificity) and 95% positive percent agreement (estimated sensitivity), when using RT-PCR as a reference. This was true for samples of asymptomatic individuals with a Ct ≤ 30.
‘…starting to measure viral protein loads in thousands of COVID-19 patients using mass spectrometry could help the technique to mature into a reliable approach to add to the pandemic readiness toolbox; in this effort, the work by Hober et al. will help revolutionize the way mass spectrometry is approached in the clinic…’ said Bart Van Puyvelde and Maarten Dhaenens.
In summary, the results presented by Hober and colleagues suggest that a scalable analytical method based on LC-MRM/MS could be used to detect SARS-CoV-2 with a high level of accuracy. The method has several advantages; high levels of sensitivity, reliability, and robustness, as well as a similar turn-around times compared to RT-PCR. Additionally, LC-MRM/MS instrumentation has been shown to reliably quantify absolute protein concentrations. Though the instrumentation is high cost, over time it can be a cost-effective complement to currently used diagnostic tools. In the future, detection methods based on LC-MRM/MS may be an essential part of future pandemic preparedness, not just for SARS-CoV-2 but also other viruses, and be valuable complements to current virus detection technologies.
The article by Hober et al., titled “Rapid and sensitive detection of SARS-CoV-2 infection using quantitative peptide enrichment LC-MS analysis” was published open access online in eLife on 8th November 2021 (Accepted November 4th 2021) under licence CC BY 4.0. The researchers have shared the mass spectrometry data on ProteomeXchange, and the ID for this dataset is PXD026366. In addition, the proteomics data have been deposited to Panorama Public, which allows for access to raw files and for integrating peak areas from TargetLynx XS, as well as visualization of all LC-MRM/MS chromatograms.
This work had no external funding.
Data
- The ProteomeXchange ID for the dataset is PXD026366
- Proteomics data have been deposited to Panorama Public
Article
DOI: 10.7554/eLife.70843
Hober, H., Tran-Minh, K. H., Foley, D., McDonald, T., Vissers, J. P C., Pattison, R., Ferries, S., Hermansson, S., Betner, I., Uhlén, M., Razavi, M., Yip, R., Pope, M. E., Pearson, T. W., Andersson, L. N., Bartlett, A., Calton, L., Alm, J. J., Engstrand, L. & Edfors F. Rapid and sensitive detection of SARS-CoV-2 infection using quantitative peptide enrichment LC-MS analysis. eLife 10:e70843 (2021).